timeSeries Class Library
- Date:
- April 2005
- Introduction
- Getting Started -- A quick tour of the timeSeries classes
- Example: Analysis of the NADS/CAMP data
- Advanced Topic: Data Segmentation
- Commnd Line Application Framework
- System Requirements
- Installation
- Debugging Tips
Introduction
Data collected from real-time systems is often acquired at uniform time intervals. This is true for both simulators like VIRTTEX and for vehicle or other hardware data acquisition systems. The goal of the timeSeries class library is to make data reduction tasks on this time-ordered or time series data easier and more efficient. Although it was written specifically to help reduce typical simulator or vehicle data sets, the classes are quite general and can be used on any time series data regardless of origin.
The timeSeries classes use features of modern C++ (classes, operator overloading, templates, exceptions and the Standard Template Library or STL) to encapsulate time-ordered data in way that allows many common operations to be vectorized.
Vectorization alone leads to a large reduction in the amount of programming effort needed for many data reduction tasks. But the timeSeries library provides other features as well:
- Flexible input format for time-ordered binary data
- The ability to handle data files with heterogenous data types
- Ability to associate text labels with data items
- A growing (read: small right now) library of statistical and mathematical functions
- A simple mechanism for collecting the results of an analysis (mesaures)
- A powerful data searching mechanism that makes it simple to locate "interesting" places in the time ordered data.
- Flexible methods for segmenting data when multiple performance intervals are contained within one data file.
- Careful attention to efficiency for both execution speed and memory management. The timeSeries class library has been designed to operate on real-world data sets that may be 10's or 100's of megabytes in size for each experimental run and where thousands of such runs may need to be reduced.
Why write a new class library when data analysis products like Matlab already provide a set of vector routines for processing data? The primary motivation was efficiency. Getting data into Matlab is often a slow and inefficient process. Matlab itself is notoriously inefficient about memory management. And Matlab m-files can be agonizingly slow for those parts of an analysis project that cannot be easily vectorized.
Getting Started -- A quick tour of the timeSeries classes
Here I'll try to give a simple tutorial on the use of the timeSeries classes. I assume that you have some familiarity with C++ concepts and with templates in particular, but I'll explain some of the details as needed.
Data Input
We need to start by getting data loaded and ready for analysis. The timeSeries classes assume that data is segregated into individual files and that each file is an experimental run that needs to be analyzed. Each data file contains data sampled at a uniform interval. An experimental data set may consist of many such data files. For most analyses the sampling interval will be the same for all the runs that make up a data set but this is not required. What is necessary is that the sampling interval be constant within a given data file.
Data Format Definition
The first task is to define the format of the data. Currently only binary data is supported, but I expect this to change in the future. For now, I'll assume you have binary data and some description of the contents of the file. The binary data must be record-oriented, that is, it must consist of all the data recorded at time t0, followed by all the data recorded at t0+dt and so on. No provision is made for a binary header of any kind.
It turns out that this describes just about all the real-time data you are likely to encounter. If that turns out to be too restrictive in the future, I may add the ability to define additional structure.
VIRTTEX data files are of exactly this form. In fact, the VIRTTEX files are very simple in that they contain one and only one data type: float. Because this format is so simple and because we analyze them most often, the timeSeries library can directly read a VIRTTEX ".hdr" file and determine the structure of a file.
For the timeSeries library, the format of a data file is encapsulated in the dataFrame class. A dataFrame is constructed from a pre-existing file, which can be a VIRTTEX ".hdr" file:
dataFrame hmi04Frame("/Users/jgreenb2/hmi04/mb_08_20d.hdr");
This code creates a dataFrame object called hmi04Frame and uses a standard ".hdr" file to define the fields.
But what if the data file didn't come from VIRTTEX? The timeSeries classes can handle formats that are more general than the VIRTTEX format. For example, the data file can contain data of types char, short, int, float or double. The data can also be grouped together into arrays. In this case we create a dataFrame definition file or ".dfm" file to define the data. The dfm format is simple and resembles a list of C-style declarators:
float CFS_Accelerator_Pedal_Position; short CFS_Auto_Transmission_Mode; float CFS_Brake_Pedal_Force; float CFS_Steering_Wheel_Angle; float CFS_Steering_Wheel_Angle_Rate; float CFS_Steering_Wheel_Torque; short CIS_Cruise_Control; float MIF_Head_Point_Angular_Velocities[3]; float MIF_Head_Point_Specific_Forces[3]; float MTS_Head_Point_Specific_Forces[3]; float MTS_Head_Point_Angular_Velocities[3]; float SCC_Lane_Deviation[4]; int SCC_DynObj_DataSize; int SCC_DynObj_CvedId[20]; int SCC_DynObj_SolId[20]; char SCC_DynObj_Name[640]; float SCC_DynObj_Pos[60]; float SCC_DynObj_Heading[20]; float SCC_DynObj_RollPitch[40]; int SCC_DynObj_AudioVisualState[20]; float SCC_DynObj_Vel[20]; float SCC_OwnVehToLeadObjDist; float VDS_Chassis_CG_Accel[2]; float VDS_Chassis_CG_Ang_Vel[3]; float VDS_Chassis_CG_Orient[3]; float VDS_Chassis_CG_Position[3]; float VDS_Steering_Torque_Backdrive;
Each line consists of one of the allowable data types followed by the name of the data item and a semicolon. Optionally, there can be an array size declaration. Array groupings mean that this data should be logically treated as an array, but it is not strictly necessary to use them to define a data layout. For example, this array declaration:
float MIF_Head_Point_Angular_Velocities[3];
could be replaced with the following three declarations without changing the layout of the data file:
float MIF_Head_Point_Angular_Velocities_1; float MIF_Head_Point_Angular_Velocities_2; float MIF_Head_Point_Angular_Velocities_3;
I wouldn't advise trying that for the SCC_DynObj_Name array! And there's another reason to use array declarators. We'll see later on that different types of data objects are created from an array (timeSeriesArray) and from a scalar (timeSeries).
Whether you have a ".dfm" file or a ".hdr" file to define your data the procedure for getting this information into your program is the same: construct a dataFrame object with the definition file as an initializer:
string framePath="/Users/jgreenb2/campAnalysis/analysis/nadsCampFrame.dfm"; dataFrame nadsCampFrame(framePath);
Reading the Data File
Now that the hard work is done, reading the file is easy. A file is represented as a dataFile object. Declaring a dataFile with the appropriate arguments automatically loads the contents of the data file into memory:
dataFile data(dataFileName,nadsCampFrame,Ts,dataFile::LittleEndian);
The first argument is the name of the data file, the second argument is a dataFrame that defines the format. The third agrument is the sampling interval in seconds. The final argument is a flag that defines the byte ordering of the data ( LittleEndian or BigEndian ) depending on the type of machine that originally created the data.
Accessing Your Data: creating timeSeries objects
Now that the data has been read in, how do you work with it? The dataFile class provides no methods for accessing the data. To gain access to the data you need to create timeSeries and/or timeSeriesArray objects. Since the ".dfm" file example from the last section is much richer in content than the VIRTTEX example, I'll use it for the rest of what follows.
To gain access to scalar data:
timeSeries<float> throttle("CFS_Accelerator_Pedal_Position", data);
Here we are declaring a scalar timeSeries object called throttle. The variable throttle will be populated by the corresponding field named "CFS_Accelerator_Pedal_Position" in the .dfm file. We know that throttle is a scalar so a timeSeries as opposed to a timeSeriesArray is the right way to represent the data. We also know that throttle is of data type float so we use C++ template syntax to declare our timeSeries. timeSeries like all template classes is really a whole set of seperate classes whos final definition depend on the value of the template parameter included between the angle brackets. So a timeSeries<float> object is just a timeSeries that is designed to work with float data. The arguments to the constructor are simply the name of the data item (from the .dfm or .hdr file) and a dataFile object that contains the data we are working with.
If you look at older code (and some of the example programs in this documentation) you will see a more complicated declaration for timeSeries objects. You don't need to use this older method in your code but in case you run across it, I'll explain how it works. The older declaration for a variable like throttle would look like this:
timeSeries<float> throttle(nadsCampFrame.findName("CFS_Accelerator_Pedal_Position"),data);
That looks a bit intimidating, but it's really very simple if we break it down. Once again, we are declaring a timeSeries object called throttle. In this form of the constructor, the first argument is a dataDef or data definition object. We need that because we need to specify exactly what part of the data file really corresponds to the throttle data. Our dataFrame variable nadsCampFrame contains all of the data definitions for the entire file so all we have to do is find the right one. We do that by using the findName member function of the dataFrame class.
The second argument is just the dataFile object that we created from the data file.
In new code you should use the first form of the declaration. Current versions of timeSeries library essentially call findName internally to find out where in the data record the requested variable is located. It's simpler and clearer and there is no advantage to using the older, more complex constructor.
If we want to access array data we can do it in a very similar way:
timeSeriesArray<float> omega("VDS_Chassis_CG_Ang_Vel",data);
The meaning here should be clear by analogy with the scalar case.
Every item from a data file that you want to use in the analysis must be represented by a timeSeries or timeSeriesArray object before you can access it.
Basic operations on timeSeries and timeSeriesArray objects
Now we're getting to the heart of the matter. A number of operations exist that allow these objects to be treated as if they were simple types. In particular, you can perform basic aritmetic:
timeSeries<float> swavel("CFS_Steering_Wheel_Angle_Rate",data); timeSeries<float> swa("CFS_Steering_Wheel_Angle",data); timeSeries<float> swaDeg = swa * 57.3; timeSeries<float> sum = swavel + swa; timeSeries<float> weightedSum = 3.0*swavel + 5.0*swa; timeSeries<float> swa2 = swa*swa;
In each of the above example, the arithmetic operations occur on every one of the data samples that make up the timeSeries object. So these operations are vectorized in that sense. Be aware that all of the arithmetic operators are defined to act element-by-element on the objects. So an expression like swa*swa creates a timeSeries where every element is the square of the corresponding element in swa.
Notice also, that you can create new timeSeries objects from old ones just by declaring them and using the ordinary "=" syntax.
You won't always want to access the entire timeSeries or timeSeriesArray object at one time. Sometimes you want a specific data sample. You can do this using ordinary C syntax:
float swa_sample = swa[2341];
The timeSeriesArray objects need 2 indices since they represent more than one data item at each data sample:
timeSeriesArray<float> omega("VDS_Chassis_CG_Ang_Vel",data); float omega_sample = omega[1][2341];
so omega_sample is the second element of omega at the 2,341st data sample.
Sometimes data sample number isn't as useful as time for locating data. You can use time as an index by substituting () for [] when writing the expression:
float swa_time = swa(23.21); float omega_time = omega[1](23.21);
If there is no data sample that corresponds to exactly 23.21 seconds, linear interpolation is used.
The timeSeries library also allows for the analysis of sections of the data called ranges. A range is created using the range member function:
timeSeries<float>swa_range = swa.range(15, 2500); timeSeries<float>swa_trange = swa.range(9.1, 12.2);
The variable swa_range contains swa values from data sample 15 to sample 2500 inclusive. The variable swa_trange contains swa values starting at the closest data sample to 9.1 seconds and continuing up to 12.2 seconds.
Ranges have the property that they inherit the time and data sample numbering from the objects that create them. This means that swa[20] is the same point as swa_range[20]. It also means that swa_range[14] is out of range and generates an error!
You can create a timeSeries from a timeSeriesArray too:
timeSeries<float>SVaccel = cgaccel.element(0);
This extracts the zeroth element of the cgaccel array and places it in a new timeSeries object.
More Mathemtical Operations
You are not limited to simple arithmetic when dealing with timeSeries objects:
// central difference timeSeries<float>POVaccel = POVspeed_filt.differentiate(); // trapezoidal integration timeSeries<float>SVspeed = SVaccel.integrate();
There are some operations that take a timeSeries object and return a scalar:
float svaMean = SVaccel.mean();
But what would happen if SVaccel were a timeSeriesArray instead of a timeSeries? In this case the mean() member function returns a set of mean values: one for every element of the timeSeriesArray. This kind of object is called a tsResult:
tsResult omegaMean = omega.mean(); for (int i=0;i<omega.get_nElem();i++) { cout << omegaMean[i] << " is the mean value of omega[" << i "]" << endl; }
As you can see omegaMean is a sequence of values, one for every element of omega. (In fact tsResult is what you get regardless of whether you invoke mean() on a timeSeries or a timeSeriesArray. It just got silently converted to a float in the first example).
Logical Comparisons
The timeSeries class (but not timeSeriesArray) supports a full range of comparison operations. So what would it mean to write an expression like (SVspeed >= POVspeed)? the timeSeries library interprets such an expression as returning a boolean value for every element of SVspeed or POVspeed (they are assumed to be the same size!). These boolean values are returned as an object called a boolResult:
boolResult faster = (SVspeed >= POVspeed);
boolResult objects themselves can logically combined using operators:
boolResult slowingDown = (SVaccel < 0.0); boolResult fasterAndSlowing = (faster && slowingDown);
The two comparisons are more often combined into a single expression:
boolResult fasterAndSlowing = (SVspeed >= POVspeed) && (SVaccel < 0.0);
Searching for Conditions
In the above example, we computed a boolResult object from the two conditions "subject vehicle
moving faster than principal other vehicle" AND "subject vehicle slowing down". If we wanted to know if that condition were true at a given data point we could simply access the boolResult variable fasterAndSlowing:
if (fasterAndSlowing[238]) { cout << "TRUE at data sample 238" << endl; }
Nice, but not that useful in most cases. The problem is that we'd like to be able to easily find out when certain conditions are true. The timeSeries classes provide just such a function. It's simply called find and it works this way:
vector<int> wherefasterAndSlowing = find((SVspeed >= POVspeed) && (SVaccel < 0.0));
The class vector is part of the STL or Standard Template Library. It's actually the base class for many of the timeSeries library objects. A vector is lot like a C array, but it has a few handy member functions like front() and back() which return the first and last values in the vector respectively. vector is a template class so we have to tell it what sort of values it should hold -- in this case we need vector<int>. That's because find returns a vector of indices that correspond to data samples that satisfy the logical condition. So if we want to know the indices of the first and last points where the conditions is true we can find out this way:
int firstIndex = wherefasterAndSlowing.front(); int firstIndex = wherefasterAndSlowing[0]; // same thing int lastIndex = wherefasterAndSlowing.back();
Experimental Measures
The object of data reduction is the computation of a small number key data items from the the time ordered data. We call such computations measures since they often correspond to key mesurements of operator or vehicle performance. Once computed, the measures need to be collected and output in a useful form.
The timeSeries library provides a class called measures that can be used for this purpose. Each computed measure has a text string associated with it that helps to identify the meaning of the item. Measures can be one of three types: int, double or string. Of course any data item that can be converted to one of these is also allowed. A measure is a type of vector quantity so that multiple measuresements can easily be collected together.
To create a measure, you declare the object and then add an entry to the measures vector. Here's how to do it:
measures myMeasures; // an empty measures object myMeasures.entry("mean sv speed",SVspeed.mean()); myMeasures.entry("Final PRNDL State", "Park"); myMeasures.entry("Subject Number", 2);
Each of these statements adds another item to the measures object. To write out just the labels as a tab-delimited text file use the print_labels function:
ofstream headerFile("mymeas.txt", ios::out);
print_labels(headerFile,myMeasures);
headerFile.close();
To write out the measures themselves you can use the output operator << to write the values to an output stream:
ofstream mFile("mymeas.dat", ios::out);
mFile << myMeasures;
mFile.close();
Once again, the values are written as tab-delimited text.
Example: Analysis of the NADS/CAMP data
I've included the actual analysis program used to process 4025 data files generated by the National Advanced Driving Simulator as a (mostly) cross-referenced example. Click on the examples link at the top of this page
Advanced Topic: Data Segmentation
The NADS/CAMP example program has one important simplification: each data file contains one and only one experimental condtion. This is not the case when many experimental treatments are presented to a test subject during the course of a single data collection run. When many conditions are present, the data reduction program needs to be concerned with breaking up the data into multiple sections that each may need to be analyzed seperately.
Experimenter's frequently include discrete variables or flags in the data stream for the express purpose of segmenting the data. In the timeSeries library, we use the stateFlag object to represent such variables.
Overview of stateFlags, states and transitions
A stateFlag is simply an integer timeSeries object. It inherits all of the attributes of a timeSeries<int> and adds a few specific functions that are useful for data segmentation. A stateflag is constructed from an existing timeSeries object:
timeSeries<float> rawCond("COND",data); stateFlag expCondition(rawCond);
Notice that the base object rawCond is a float but that stateFlag objects are always int objects. This is the common condition in VIRTTEX data since the VIRTTEX data file format stores only float data. The stateFlag constructor converts the data to int when creating the stateFlag. This can make creating stateFlag objects considerably more expensive than just referencing exisitng data. The entire underlying timeSeries must be copied and converted to an int when a stateFlag is constructed.
If the underlying data is of integer type the data conversion process can be bypassed and the stateFlag can be created directly from the dataFile:
stateFlag expCondition("INT_COND",data);
However, if INT_COND does not correspond to an integer field this will result in a BadType exception at run time.
Several types of stateFlag objects are commonly found in experimental data. The simplest form is a binary variable that is equal to zero except when a specific condition is true.
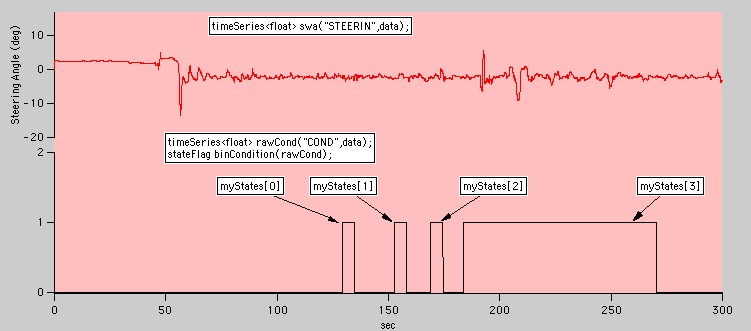
A simple binary flag
stateFlag and its associated classes (transition, state and stateFlag::iterator) provide methods for using this type of information to easily segment data. For example, the four states defined by the stateFlag binCondition in the graph above can be found using the findStates member function:
vector<state> myStates = binCondition.findStates();
It certainly looks like the steering angle response in the fourth state differs from the other three states. We can focus on just this state by using a state variable to define a range:
timeSeries<float> swaInState4 = swa.range(myStates[3]);
Iterating Over States: Binary Data
More often, we need to iterate over all the experimental conditions and compute measures. There are many (perhaps too many!) ways to do this, depending on what is needed. Perhaps the simplest way is to use the findStates function that we have already seen:
vector<state> myStates = binCondition.findStates(); for (int i=0;i<myStates.size();++i) { computeMeasure(swa.range(myStates[i])); }
By now, it should be clear that a state is akin to a range: it defines a temporal region, bounded by two transitions. In some cases, we don't actually need to know the entire state, we just want to know where the transitions occur. There are two functions for finding these:
vector<int> risingEdgeLocations = binCondition.rising(); vector<int> fallingEdgeLocations = binCondition.falling();
The rising and falling member functions individually do less work than findStates since they only bother to locate the starting or ending point of the states.
Iterating Over States: Polytomous Data
Data flags are often not just binary values. To simplify data collection and to cut down on the number of channels, a stateFlag may take on one of several discrete values that encode information about the type of the state.
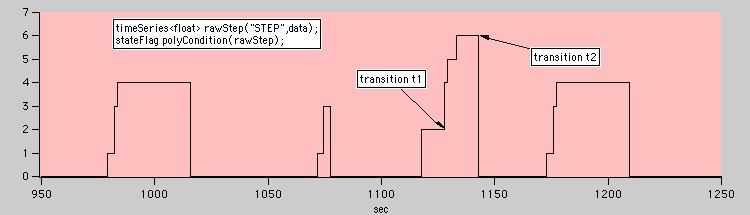
A Polytomous flag
By default the findStates member function looks for states bounded by any rising edge transition followed by any falling edge transition. But for polytomous data we may want to be more specific. We can instruct findStates to use well-defined transitions when looking for states:
transition t1(2,4); transition t2(6,0); vector<state> myStates = polyCondition.findStates(t1,t2);
Iterating Over Edges
The preceding methods are adequate for many cases. However, the findStates function is designed to find all of the states contained in its associated stateFlag object. If we are going to need to find them all that's fine. But if we only need to find the next transition, using findStates wastes a lot of time. For this reason, the stateFlag class provides iterators for directly iterating over rising or falling edges.
A stateFlag iterator is a template class that must be instantiated with one of three comparator classes: rising, falling or change. For example, if we wanted to find the location and value of polyCondition at its first three rising edges we could compute it this way:
stateFlag::iterator<rising> nextEdge=polyCondition.begin<rising>(); for (int i=0;i<3;++i) { cout << "rising edge at index: " << nextEdge.index(); cout << "value = " << *nextEdge << endl; ++nextEdge; }
This takes a little explaining (I did put this in the advanced section!). First, an iterator is a general STL concept. It's a generalization of a simple pointer and has many of the same properties. In particular, iterators can be dereferenced using the * operator to give the value of their associated object at the current iterator position. They can also be incremented and decremented. stateFlag objects always provide member functions begin and end which return iterators pointing to the first edge and just past the last edge.
So now let's look more closely at the above example line-by-line:
stateFlag::iterator<rising> nextEdge=polyCondition.begin<rising>();
There are many types of iterators and the stateFlag iterator exists only in the stateFlag namespace, hence the stateFlag::iterator notation. These iterators are also template classes and here we want to specify that we iterating over rising edges. In the same line, we initialize nextEdge using the begin member function of the polyCondition object.
polyCondition.begin<rising>() returns a rising edge iterator that points to the first rising edge in the polyCondition stateFlag object. Let's assume for the purposes of this example that the first rising edge is the one that occurs in the graph just after 975 seconds.
for (int i=0;i<3;++i) { cout << "rising edge at index: " << nextEdge.index();
The goal was to find 3 edges so that's how the loop is set up. nextEdge is alread pointing to the first rising edge, that's how we initialized it. To find out where that edge is located we use the index() member function which returns the integer index corresponding to the current iterator position.
cout << "value = " << *nextEdge << endl;
Now we want to print the value at the edge location. We could just reference polyCondition[nextEdge.index()] but that's more work than needed. Remember that a iterator is a generalization of a pointer. The expression *nextEdge returns the value of polyCondition at the current iterator location.
++nextEdge; }
We increment the nextEdge iterator using ++ which advances to the next rising edge. In the C language it would be more common to see this written as nextEdge++ rather than ++nextEdge. Both expressions are valid and have the same effect in our example. But there's an efficiency difference between them. A post-increment expression like nextEdge++ has a value that is equal to nextEdge before it is incremented. That means that the compiler must make a copy of nextEdge before it performs the increment operation. By contrast the pre-increment expression ++nextEdge returns the value of nextEdge after the increment operation so no copy needs to be created. The overhead isn't much in this case since these objects are small, but as a point of style you will rarely see a post-increment operator used in C++ unless the return value is explicitly used.
stateFlag iterators can also be compared. The result of an expression like (i1 < i2) where i1 and i2 are both iterators is true if the index associated with i1 is less then the index associated with i2. This makes it easy to write loops that range over a data set. The following example prints the location of every falling edge iterator contained in polyCondition along with the value at the edge:
stateFlag::iterator<falling> nextEdge = polyCondition.begin<falling>(); stateFlag::iterator<falling> lastEdge = polyCondition.end<falling>(); while (nextEdge < lastEdge) { cout << "falling edge at index: " << nextEdge.index(); cout << " value= " << *nextEdge++ << endl; }
In this case the expression *nextEdge++ is deliberately a post-increment because we need to dereference nextEdge prior to advancing it. More importantly, you should be aware that the begin(), end() and increment/decrement functions on stateFlags operate in linear time in the current implementation. The end() function in particular can be slow if the last edge is far from the end of the stateFlag object.
Discrete variables that do not define states
Finally, there are often cases where an experimenter creates a data channel that simply transitions to a discrete integer value whenever a new condition occurs.
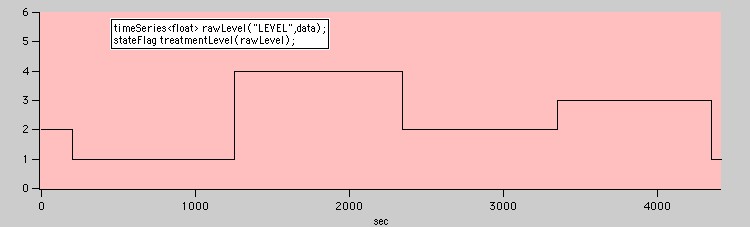
A discrete variable that does not define states!
Although the intention may be to define 'states' that correspond to the different values of the treatmentLevels object, a call like:
vector<state> myStates = treatmentLevels.findStates();
Will not return the desired result! The reason is that findStates needs a consistent definition of the begining and ending transitions associated with the states. If you call findStates without arguments it defaults to these:
findStates(transition::risingEdge, transition::fallingEdge);
Where transition::risingEdge and transition::fallingEdge are constant transition objects that match rising and falling edges respectively. In the current example that means that findStates will return only two states, not the four states you may be expecting.
If you wish to find all of these states there are many possiblities. You can make a second call to findStates and explicitly look for falling edges followed by rising edges:
findStates(transition::fallingEdge, transition::risingEdge);
However, you'll still need to splice the two resulting vectors together. Alternatively you can assemble the states directly using iterators and the state object constructor:
stateFlag::iterator<change> nextEdge=treatmentLevels.begin<change>();; stateFlag::iterator<change> lastEdge = treatmentLevels.end<change>(); stateFlag::iterator<change> prevEdge; vector<state> myStates; while (nextEdge < lastEdge) { prevEdge = nextEdge; state s(prevEdge.index(),(++nextEdge).index()); myStates.push_back(s); }
The examples section includes a program called hmi04.cpp that uses stateFlag objects extensively to segment an analysis.
Commnd Line Application Framework
A simple application framework is included with the timeSeries classes. The framework makes it easy to write a command-line application capable of processing thousands of data files with a single statement. The framework class is called tsCmdLineApp and the documentation for this class contains instructions on how to use it.
System Requirements
The timeSeries libraries require a computer with an ISO C++ compiler and full support for the C++ Standard Template Library (STL). Currently, there is support for GNU C++ 3.4 or greater and Microsoft Visual C++ 7.1 or greater. Earlier versions of these compilers will not work. Other compilers may work but have not been tested. To the extent possible, the code is ISO compliant but I have no way to guarantee this. If you choose to use a Microsoft compiler the optimizing "Professional" versions are required in order to achieve acceptable performance.
The code is processor-independent and will automatically adapt to big-endian or little-endian architectures. The libraries are also largely operating system independent although the example programs do include a facility to correctly handle Windows "shortcuts". For convenience, the base code to support this is contained in the timeSeries library.
The main issue when selecting a target computer is memory since the design of the timeSeries libraries relies on loading an entire data file into memory at once. The example programs show how to handle thousands of such files in a single run. Experience has shown that there are no inherent memory leaks in the libraries so a correctly written application can process many files using only enough memory to hold the largest data file plus any working storage. As a practical matter, main memory that is 1.5x the size of the largest data file to be processed will be adequate in most cases.
A less important issue is the command-language ability of the target system. The example programs are all command-line driven -- no GUI is provided. This is often the simplest and most effective way to deal with the type of data reduction required for vehicle or simulator experiements because working with hundreds or thousands of files is easier when a good command-line interpreter is available. Unix systems excel at this, of course. Under Windows, the example programs support wildcard matching and the use of Windows shortcuts in the DOS window. Additional file manipulation can be performed under Windows using Perl if necessary or by using Cygwin.
Finally, the timeSeries libraries rely on smart pointers from the Boost libraries for memory management. These libraries must be properly installed on the target system.
Installation
The Boost Libraries
Boost is an open-source project to extend the STL with additional capabilties. The only Boost facility used by the timeSeries library are smart pointers. Along with many other Boost extensions, the Boost smart pointer library is in the process of being adopted into the ISO Standard Template Library. Until that time, however, smart pointers must be installed separately. Current versions of the library can always be found at http://www.boost.org. Installing Boost is a two-step process. First you must download (or build) the "Jam" tool which is used by Boost in place of the usual "Make" facility.
Once Jam is installed you can download the source for the Boost libraries and build the distribution. Detailed instructions are given at http://www.boost.org/more/getting_started.html .
The Subversion Source Code Control System
The timeSeries source code resides on osprey.srl.ford.com. You access the source using the Subversion source code control system http://subversion.tigris.org/. Subversion is much like CVS but contains many improvements. Subversion is robust, cross-platform and free. The Subversion website includes an HTML version of the book "Version Control with Subversion". The book is also available in printed form at bookstores.
To gain access to the Subversion source respository first contact me (jgreenb2@ford.com) and I'll grant you access. Then you will need to download the latest Subversion toolset. There are pre-built executables for Windows, Linux, Mac OS X and other systems.
Once SVN is installed (and once you have requested access) you can examine the contents of the Repository like this:
C:>svn ls svn://osprey.srl.ford.com campAnalysis/ camp_virttex/ hmi04/ timeSeries/
The libraries themselves are in the timeSeries/ directory. You should not need to modify this directory. To check out an example program use the svn checkout command:
C:>svn checkout svn://osprey.srl.ford.com/hmi04 A hmi04\hmi04.1 A hmi04\hmi04.cpp A hmi04\hmi04.xcode A hmi04\hmi04.xcode\jgreenb2.mode1 A hmi04\hmi04.xcode\jgreenb2.pbxuser A hmi04\hmi04.xcode\project.pbxproj A hmi04\hmi04_utilities.h A hmi04\hmi04.vcproj A hmi04\hmi04.h A hmi04\hmi04.xcodeproj A hmi04\hmi04.xcodeproj\jgreenb2.mode1 A hmi04\hmi04.xcodeproj\jgreenb2.pbxuser A hmi04\hmi04.xcodeproj\project.pbxproj A hmi04\hmi04_utilities.cpp A hmi04\vsim_test.cpp U hmi04 Fetching external item into 'hmi04\timeSeries' A hmi04\timeSeries\lookupTables A hmi04\timeSeries\lookupTables\lookupTable.cpp A hmi04\timeSeries\lookupTables\lookupTable.h A hmi04\timeSeries\timeSeries Classes A hmi04\timeSeries\timeSeries Classes\tsVal.h A hmi04\timeSeries\timeSeries Classes\boolResult.h A hmi04\timeSeries\timeSeries Classes\tsAnalysis.h A hmi04\timeSeries\timeSeries Classes\timeSeriesOverview.h A hmi04\timeSeries\timeSeries Classes\timeSeriesArray.cpp A hmi04\timeSeries\timeSeries Classes\timeSeries.cpp A hmi04\timeSeries\timeSeries Classes\dataFile.cpp A hmi04\timeSeries\timeSeries Classes\stateFlag.cpp A hmi04\timeSeries\timeSeries Classes\timeSeriesArray.h A hmi04\timeSeries\timeSeries Classes\timeSeries.h A hmi04\timeSeries\timeSeries Classes\dataFile.h A hmi04\timeSeries\timeSeries Classes\stateFlag.h A hmi04\timeSeries\timeSeries Classes\tsBase.cpp A hmi04\timeSeries\timeSeries Classes\measures.cpp A hmi04\timeSeries\timeSeries Classes\dataFrame.cpp A hmi04\timeSeries\timeSeries Classes\state.h A hmi04\timeSeries\timeSeries Classes\tsResult.cpp A hmi04\timeSeries\timeSeries Classes\tsBase.h A hmi04\timeSeries\timeSeries Classes\measures.h A hmi04\timeSeries\timeSeries Classes\dataFrame.h A hmi04\timeSeries\timeSeries Classes\boolResult.cpp A hmi04\timeSeries\timeSeries Classes\tsAnalysis.cpp A hmi04\timeSeries\timeSeries Classes\tsResult.h A hmi04\timeSeries\timeSeries Classes\tsExceptions.h A hmi04\timeSeries\filters A hmi04\timeSeries\filters\filter.c A hmi04\timeSeries\filters\filter.h A hmi04\timeSeries\Documentation A hmi04\timeSeries\Documentation\Doxyfile A hmi04\timeSeries\Documentation\images A hmi04\timeSeries\Documentation\images\stateFlag_graphs.pxp A hmi04\timeSeries\Documentation\images\timeseries1.JPG A hmi04\timeSeries\Documentation\images\timeseries2.JPG A hmi04\timeSeries\Documentation\images\timeseries3.JPG A hmi04\timeSeries\Windows A hmi04\timeSeries\Windows\windowsMain.cpp A hmi04\timeSeries\Windows\Windows Header Files A hmi04\timeSeries\Windows\Windows Header Files\stdafx.h A hmi04\timeSeries\Windows\Windows Header Files\windowsMain.h A hmi04\timeSeries\Windows\Resource Files A hmi04\timeSeries\Windows\Resource Files\campAnalysis.rc A hmi04\timeSeries\Windows\Resource Files\resource.h A hmi04\timeSeries\Windows\stdafx.cpp A hmi04\timeSeries\Windows\shortcuts A hmi04\timeSeries\Windows\shortcuts\resolveShortCut.h A hmi04\timeSeries\Windows\shortcuts\resolveShortCut.cpp A hmi04\timeSeries\utilities A hmi04\timeSeries\utilities\tclap A hmi04\timeSeries\utilities\tclap\SwitchArg.h A hmi04\timeSeries\utilities\tclap\CmdLineOutput.h A hmi04\timeSeries\utilities\tclap\CmdLineInterface.h A hmi04\timeSeries\utilities\tclap\Visitor.h A hmi04\timeSeries\utilities\tclap\UnlabeledMultiArg.h A hmi04\timeSeries\utilities\tclap\IgnoreRestVisitor.h A hmi04\timeSeries\utilities\tclap\DocBookOutput.h A hmi04\timeSeries\utilities\tclap\VersionVisitor.h A hmi04\timeSeries\utilities\tclap\ValueArg.h A hmi04\timeSeries\utilities\tclap\XorHandler.h A hmi04\timeSeries\utilities\tclap\HelpVisitor.h A hmi04\timeSeries\utilities\tclap\Arg.h A hmi04\timeSeries\utilities\tclap\StdOutput.h A hmi04\timeSeries\utilities\tclap\CmdLine.h A hmi04\timeSeries\utilities\tclap\UnlabeledValueArg.h A hmi04\timeSeries\utilities\tclap\MultiArg.h A hmi04\timeSeries\utilities\tclap\ArgException.h A hmi04\timeSeries\utilities\stringtok_std.h A hmi04\timeSeries\Examples A hmi04\timeSeries\Examples\hmi04.cpp A hmi04\timeSeries\Examples\hmi04_utilities.h A hmi04\timeSeries\Examples\campAnalysis.cpp A hmi04\timeSeries\Examples\hmi04.h A hmi04\timeSeries\Examples\hmi04_utilities.cpp U hmi04\timeSeries Checked out external at revision 193. Checked out revision 193. C:>
Once this command is complete there will be a new folder named "hmi04" on the target machine:
Result of svn checkout of the hmi04 example project
Notice that this example includes a Visual Studio project that can be opened and used to develop the application. There are also xcode project files for use on Mac OS X with Xcode and GCC. The timeSeries directory contains the source code for all timeSeries objects and does not need to be edited. The source files at the top level of the folder (hmi04.cpp, etc.) constitute the application.
To start an entirely new application, base it off an existing application that is closest to what you think you'll need. Both the hmi04 and camp_virttex projects are designed to handle many VIRTTEX post-process files in a single run and might be good starting points for many applications. Even better use the tsCmdLineApp framework to quickly create your data procesing app. To use it, just create a new version in the svn Repository with the svn copy command like this:
C:>svn copy -m "making new project" svn://osprey/tsCmdLineFrameworkApp svn://osprey/myNewApp
You can then checkout "myNewApp" and work with it as your own. There is a plug-in for Visual Studio that makes using Subversion easier. It's called Ankh and can be found at http://ankhsvn.tigris.org/ . I highly recommend it but it isn't required. If you have Ankh installed then you can easily rename the source files to reflect your new work from within Visual Studio itself. Another nearly indispensible bit of freeware for Windows if you are using SVN is called TortoiseSVN. You can find it at http://tortoisesvn.tigris.org/ . If you are running SVN under Windows you really must get this!
The Subversion respository does not currently use any special protection on the timeSeries libraries. If you find a bug in the library, report it to me (jgreenb2@ford.com) but please don't commit any changes to the timeSeries classes back to the Repository. You are, of course, encouraged to commit changes in your application as often as required. In the future I may institute a more elaborate protection scheme for the Repository.
Debugging Tips
Using the timeSeries library -- or any STL-based class library brings poses some challenges for the user. Seemingly trivial syntax errors often generate dozens of lines of nearly incomprehensible messages from the compiler. And once compiled, it is often tedious to use a traditional debugger to examine the state of complex objects. These difficulties are not unique to the timeSeries libararies. They are present in any heavily templatized C++ code -- especially code that makes extensive use of STL containers.
Compilation Errors
TBD
Using a Debugger
There are two types of problems that occur when using a debugger on programs written using the timeSeries library. First, some knowledge of the implementation details of timeSeries objects is required to effectively "see inside" of them. Second, the facilities in some debuggers (particularly Microsoft Visual Studio .NET 2003) for examining STL containers are poor.
When debugging it is often useful to know the contents of a timeSeries object. Consider the following snapshot of a debugging session:
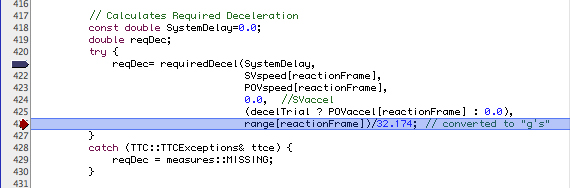
Debugging a timeSeries Application
At this point, it is reasonable to ask what is the value of POVaccel[reactionFrame]? If you just ask the debugger directly you get a surprising and not very helpful result (the example uses the GNU gdb debugger):
(gdb) p POVaccel[reactionFrame]
$8 = {
i = 34584,
j = 0,
ts = 0xbffff068
}
The problem is that an expression like POVaccel[reactionFrame] returns a tsVal object, not a scalar. During normal program execution this tsVal object will be converted to a scalar as needed, using conversion operations defined in the timeSeries library. Most debuggers, however, are not sophisticated enough to realize this. To actually access the underlying data it is necessary to know that every timeSeries or timeSeriesArray object contains a Boost smart pointer to the underlying data sequence. This pointer is simply called data and is a member of the base class for timeSeries and timeSeriesArray. Using the whatis facility of the gdb debugger we can easily see how the data member is defined:
(gdb) whatis POVaccel.data
type = shared_ptr<std::vector<float, std::allocator<float> > >
shared_ptr is the type associated with a Boost smart pointer. So POVaccel.data is a smart pointer to a vector of float. To find the answer to our original query in gdb we would type:
(gdb) p (*POVaccel.data)[reactionFrame] $1 = (float &) @0xf84bc60: -4.83007431 Current language: auto; currently c++
To get the best results with the GCC compilers use the flag '-gfull' instead of '-g' to request deugging symbols.
The debugging session above relies on two properties of the gdb debugger. First, it can properly dereference a Boost smart pointer so that an expression like *POVaccel.data actually refers a vector of float. The second is that the [] operator works with an STL vector. Unfortunately, neither of these assumptions holds for Microsoft Visual Studio .NET 2003. Visual Studio .NET 2005 is reported to be much better in this respect but I have not had the opportunity to test it.
You can still debug with VS 2003 using some workarounds. The figure below shows the same debugging session in Visual Studio.
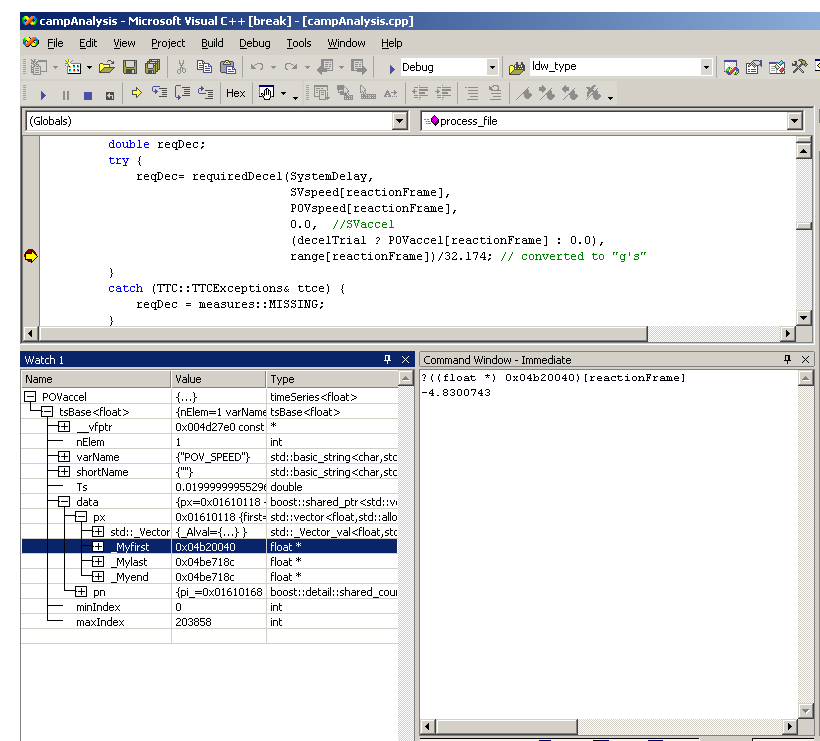
Debugging with MS Visual Studio
To obtain the value of POVaccel[reactionFrame] you must enter POVaccel into the "Watch" window. Then expand data members as shown on the left hand side of the figure until the "_Myfirst" field is revealed. The hex address in this field is the base address of the POVaccel data sequence. You can double-click this field and copy the hex address so you don't need to write it down.
In the "Command" window enter the expression:
? ((float *) 0x04b20040)[reactionFrame]
This casts the hex pointer to be a pointer to float and then uses the [] operator to index the array and return the requested data value. Messy, but not terribly hard once you get used to it. As I mentioned before this should be much improved in the newest versions of Visual Studio.
 1.6.3
1.6.3